 True Cell Simulations Inc. offers our customers advanced services with our A-Cell software and long-term experience in 4D cell simulation.
True Cell Simulations Inc. offers our customers advanced services with our A-Cell software and long-term experience in 4D cell simulation.
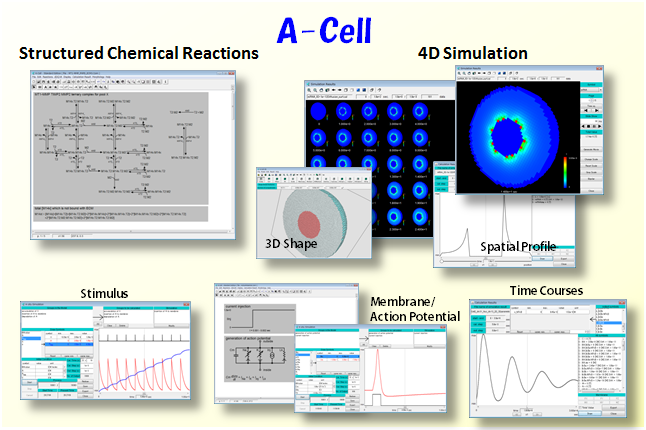
4D cell simulation is essential for revealing mechanisms of a cell. A-Cell provides an easiest environment for pathway and 4D cell simulation, and is a powerful tool both for molecular cell biologists and researchers of drug discovery.
NEWS
Dec. 23, 2017 The sixth publication of “Predictive Simulation” (“Experimental Medicine”, Yodosha, Tokyo Japan) has been released. We are introducing our predictions by stochastic simulation and their validation by wet experiments on the assembly of stress granules (Ohoshima, D., et al., PLoS Comp.Biol., e1004326, 2015).
Nov. 28, 2017 The fifth publication of “Predictive Simulation” (“Experimental Medicine”, Yodosha, Tokyo Japan) has been released. We have introduced predictions by pathway simulation and their validation by wet experiments on MAPK signaling (Bhalla, U.S., et al., Science, 1018-1023, 2002).
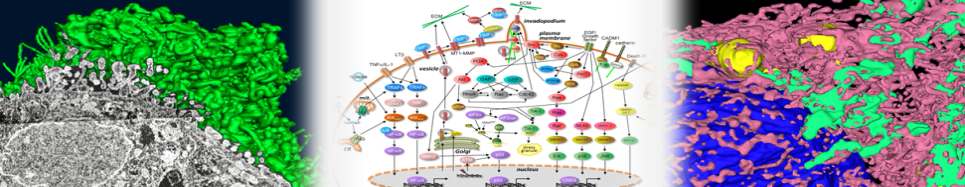
Oct. 25, 2017 The fourth publication of “Predictive Simulation” (“Experimental Medicine”, Yodosha, Tokyo Japan) has been released. We have introduced our predictions by A-Cell 4D simulation showing that oscillation frequency and persistence of a transcription factor NF-κB in the nucleus is differentially regulated by nuclear export of IκB mRNA and import of IκB protein. This is a typical example of prediction by 4D cell simulation (Ohoshima, D., et al., PLoS ONE, e0127633, 2015).
Sep. 22, 2017 The third publication of “Predictive Simulation” (“Experimental Medicine”, Yodosha, Tokyo Japan) has been released. We have introduced our predictions by A-Cell 4D simulation showing that activation of a transcription factor NF-κB in the nucleus is extensively regulated by spatial parameters including diffusion coefficients of proteins, nuclear/cytoplasmic volume ratio, nuclear transport, and the site of the protein synthesis. These are suggestive predictions for signal transduction in a cell (Ohoshima, D., et al., PLoS ONE, e109895, 2014).
Sep. 1, 2017 The second publication of “Predictive Simulation” (“Experimental Medicine”, Yodosha, Tokyo Japan) has been released. We have introduced our prediction on synaptic plasticity in neuroscience by pathway simulation of A-Cell, where strong stimuli led to long-term depression of synaptic efficacy instead of long-term potentiation. Although this prediction was counter-intuitive, its mechanism was made clear by simulations and analyses, and was confirmed by experiments (Ichikawa, K., et al., Neurocomputing, 2055-2059, 2007).
Aug. 7, 2017 A new serial publication entitled “Predictive Simulation” (“Experimental Medicine”, Yodosha, Tokyo Japan) by Kazuhisa Ichikawa、CEO of our company has started. In the first publication this month, our prediction on cancer invasion by 4D simulation of A-Cell and its confirmation by wet experiment has been introduced. Seven such examples, which are mainly based on our research, will be published monthly (Hoshino, D., et al., PLoS Comp.Biol., e1002479, 2012).
Mar. 25, 2017 YouTube video “Simulated FRAP by A-Cell” has been released. You will find A-Cell operations for constructing a model, running A-Cell simulation, and viewing simulation results for Simulated FRAP as an example of 4D cell simulation.
Oct. 1, 2016 A new eBooklet “Learn 4D Cell Simulation in 10 Pages” has been released, which is freely downloadable from our website. Differences from pathway simulation, basic concept of 4D cell simulation, its mechanisms, and an example are described in this eBooklet.
Jul. 19, 2016 A new eBooklet “Pathway Simulation by A-Cell: ERK Signal Transduction” has been released, which is freely downloadable from our website. We describe a reconstruction of a published simulation study on ERK signaling by A-Cell. We will publish a series of eBooklets on A-Cell reconstructions of simulation studies reported in journals.
Jul. 3, 2016 We have started A-Cell simulation service. We run simulations according to your experimental results and return a comparison between your experimental and our simulation results together with an A-Cell model. Visit the page of this new service through “Simulation Service“, which also appears on the top menu.
Jul. 3, 2016 We have started a free service of reconstructing published simulation by A-Cell. Open a menu “Reconstruction of published simulation by A-Cell” in the top menu “A-Cell” for detailed information.
Apr. 18, 2016 We have started selling A-Cell. Entry, Basic, and Standard Editions are purchasable on our website. Click “Buy A-Cell” menu for more information.
Apr. 18, 2016 “A-Cell Operation Manual” has been released. This manual is downloadable by A-Cell users who purchased A-Cell.
Apr. 4, 2016 A freely downloadable eBooklet “Learn Utilization of Cell Simulation in 10 Pages” has been published. This eBooklet introduces eight examples of cell simulations in molecular cell biology and drug discovery showing how collaborative research between wet experiments and in silico studies leads to unexpected and novel findings.
Mar.25, 2016 A new version of “Introduction to A-Cell” has been released.
Mar. 6, 2016 An eBooklet “Learn Pathway Simulation in 10 Pages“, which is freely downloadable from our website, has been published. The intended audience includes students and professional researchers that have expertise in experimental cell biology, pharmacology, or medical science.
Feb. 25, 2016 We will present our A-Cell software and services on cell simulation at BioAsia2016 on Mar.16, which will be held in Tokyo Mar. 15-16 2016.
Nov. 1, 2015 A document “Introduction to A-Cell” has been released describing all functions of A-Cell.
Oct. 20, 2015 We have published two book chapters: 1) Ohshima, D., et al., “Potential Roles of Spatial Parameters in the Regulation of NF-κB Oscillations, as Revealed by Computer Simulations”, pp63; 2) Ohshima, D., et al., “Stochastic Simulation of Stress Granules”, pp.77. Both are chapters in ”Protein Modifications in Pathogenic Dysregulation of Signaling”, eds. Inoue, J-I. & Takekawa, M., Springer, Tokyo, 2015, ISBN 978-4-431-55560-5, ISBN 978-4-431-55561-2 (eBook).
Oct. 14-16, 2015 We exhibited our consultation and A-Cell software at BioJapan2015, Yokohama Oct. 14-16 2015. We also presented “Qualifying Drug Discovery by Cell Simulation” at a seminar on Oct. 15.
Aug. 6, 2015 We organized a symposium entitled “Extensive Collaboration between Wet Experiment and Cell Simulation: Toward a New Era of Drug Discover” at The Institute of Medical Science, The University of Tokyo, Tokyo. In this symposium, wet and in silico researchers talked on the same topic of their collaborative research. There were 5 such sessions followed by a panel discussion, in which discussion on the essential role of cell simulation was held.
Jul. 29, 2015 Our paper “Spatio-temporal dynamics and mechanisms of stress granule assembly” has been published in PLoS Computational Biology. Cells exposed to a stress such as low oxidative condition (hypoxia) or chemicals such as arsenic acid trigger the assembly of stress granules (SGs), which are non-membranous structure of mRNA and proteins. SGs plays an important role for cell fate decision. In this paper we revealed dynamics of SG assembly in collaboration with wet experiments. We ran stochastic simulation because SGs are countable entity in the cytoplasm. (A-Cell model and stochastic simulation program are downloadable from the following button.)
Jul. 18, 2015 Our paper ”Synergistic effect of blocking cancer cell invasion revealed by computer simulations” has been accepted in Journal of Mathematical Biosciences and Engineering. We discussed potential therapeutic methods to block cancer invasion and a role of MMP-2 in the degradation of ECM proteins. (A-Cell model is downloadable from the following button.)
Jul. 4, 2015 Our paper “Regulation of NF-κB oscillation by nuclear transport: Mechanisms determining the persistency and frequency of oscillation” has been published in PLoS ONE. In the previous paper we reported that transport through nuclear membrane regulated oscillation pattern (frequency and persistency) of NF-κB (Ohshima, D., et al., PLoS ONE, 7 (2012), e46911). Here we analyzed how and why nuclear transport regulated the oscillation pattern and found that export of IκB mRNA and import of newly synthesized IκB regulated persistency and frequency independently. (A-Cell models are downloadable from the following button.)
Apr. 21, 2015 Our paper “Regulation of NF-κB oscillation by nuclear transport: Mechanisms determining the persistency and frequency of oscillation” has been accepted in PLoS ONE.
Apr. 8, 2015 Our paper “Spatio-temporal dynamics and mechanisms of stress granule assembly” has been accepted in PLoS Computational Biology.